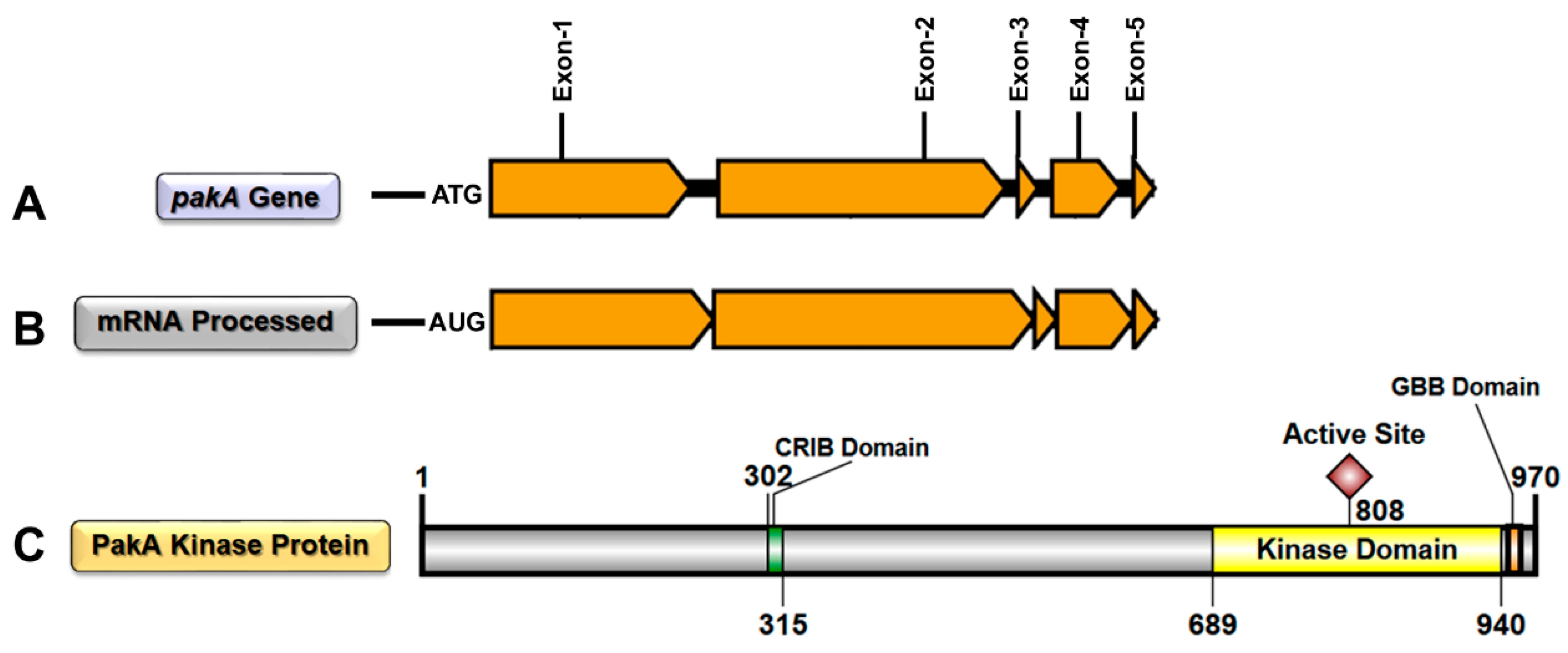
IJMS | Free Full-Text | STE20/PAKA Protein Kinase Gene Releases an Autoinhibitory Domain through Pre-mRNA Alternative Splicing in the Dermatophyte Trichophyton rubrum | HTML
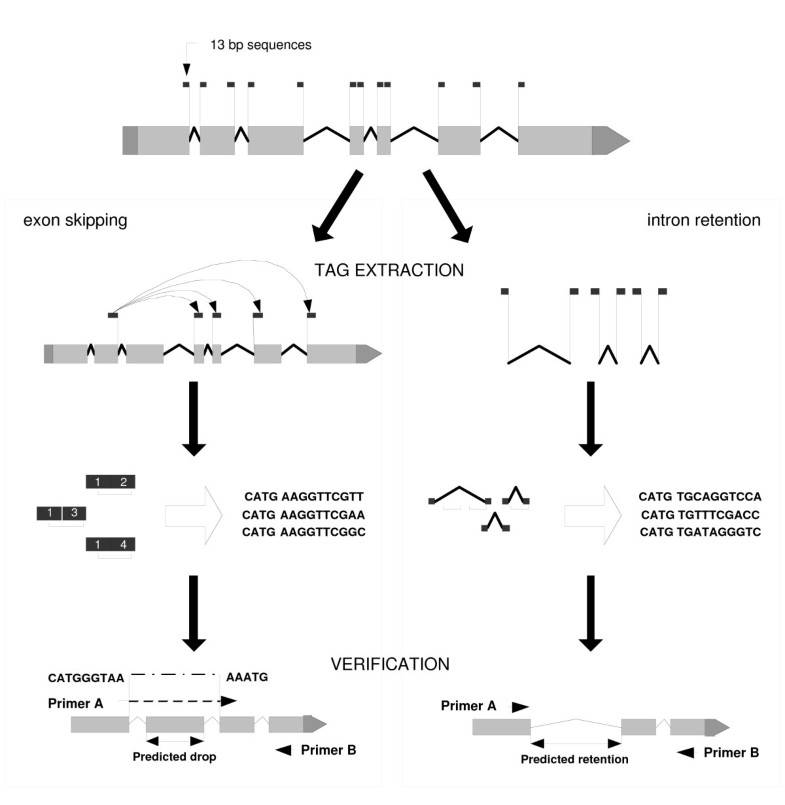
Discovery of novel alternatively spliced C. elegans transcripts by computational analysis of SAGE data | SpringerLink
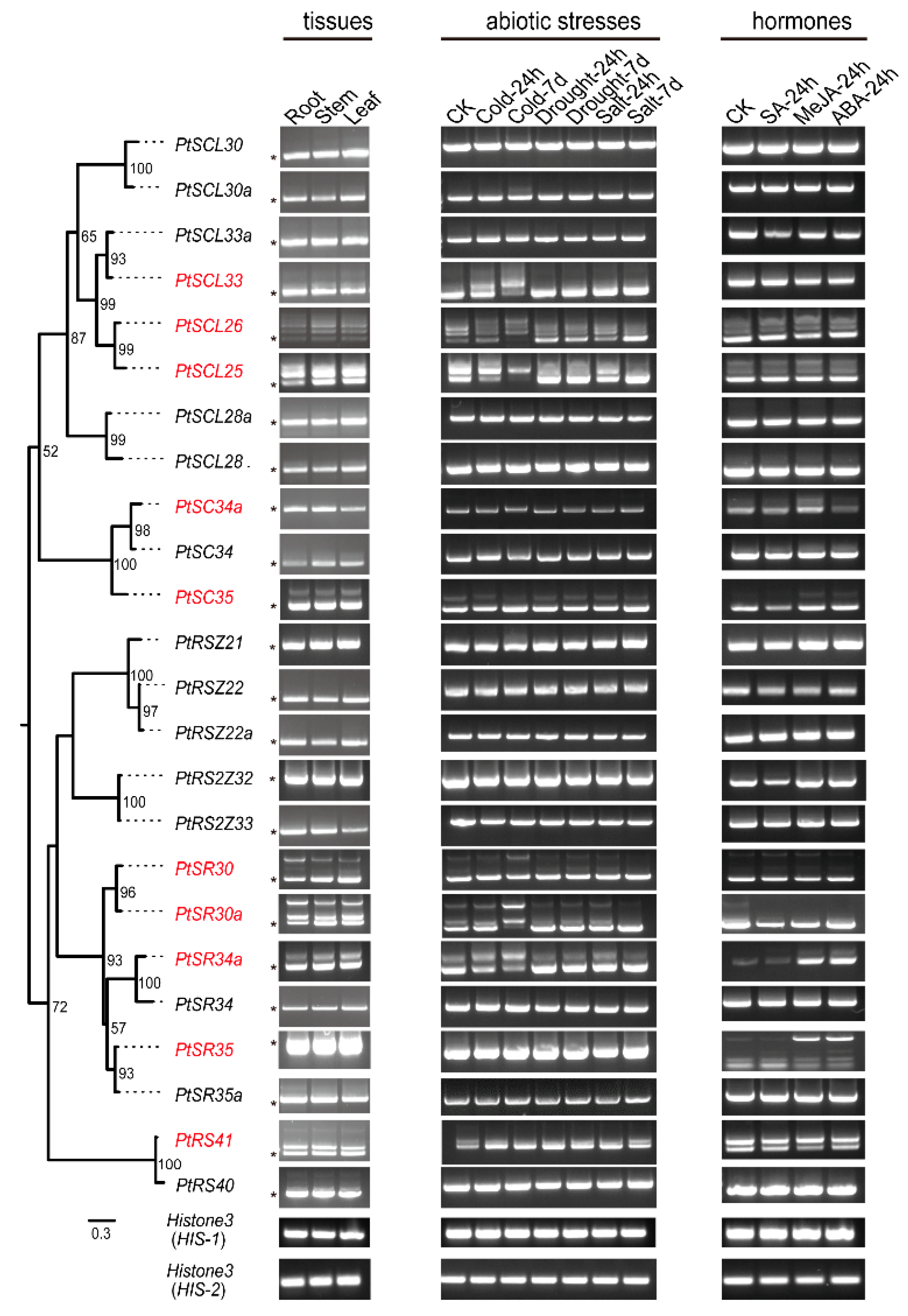
IJMS | Free Full-Text | The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa | HTML
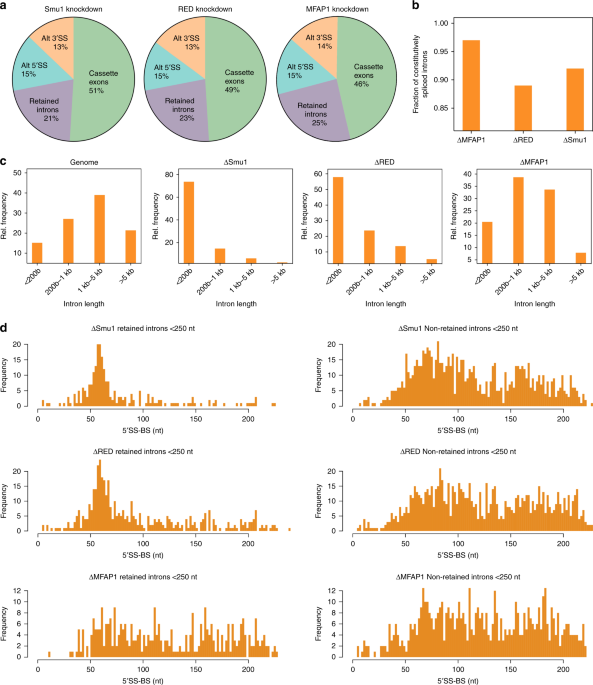
Smu1 and RED are required for activation of spliceosomal B complexes assembled on short introns | Nature Communications
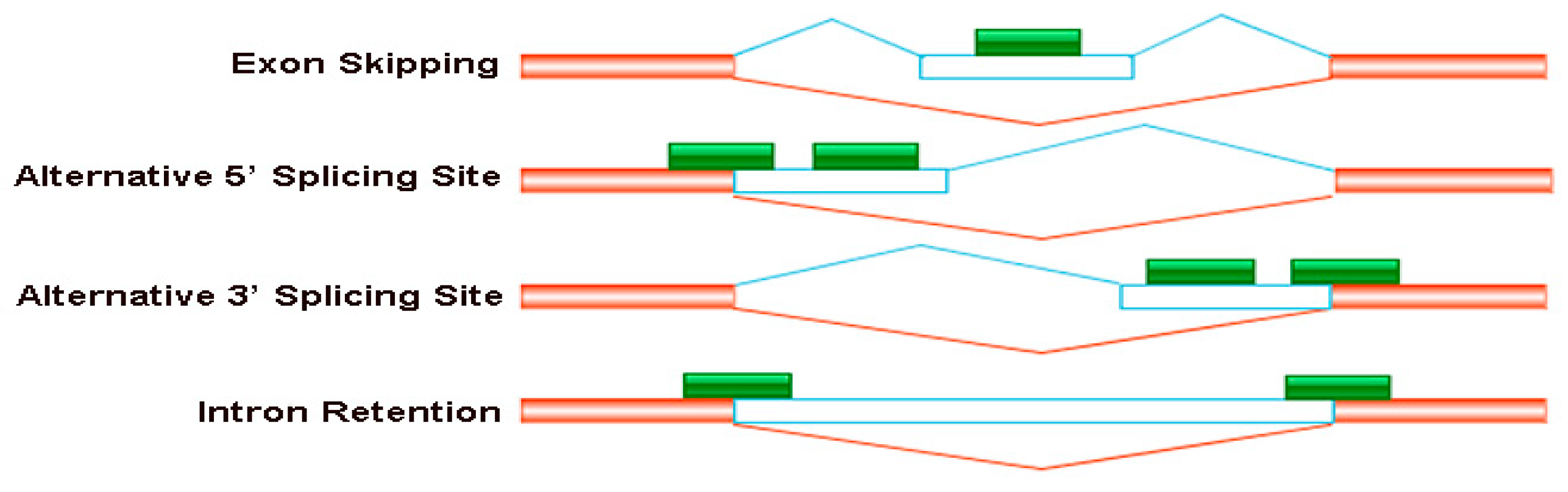
Genes | Free Full-Text | Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus | HTML
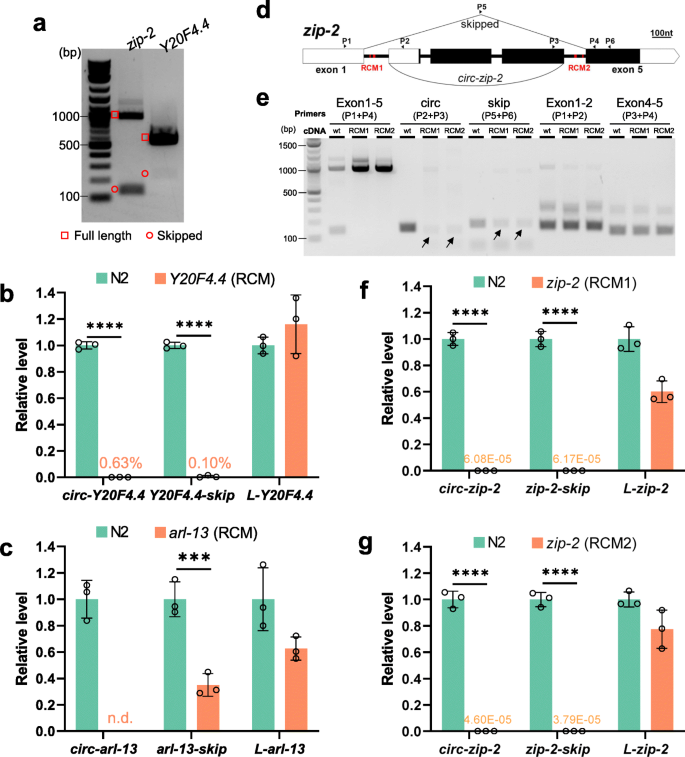
Reverse complementary matches simultaneously promote both back-splicing and exon-skipping | SpringerLink
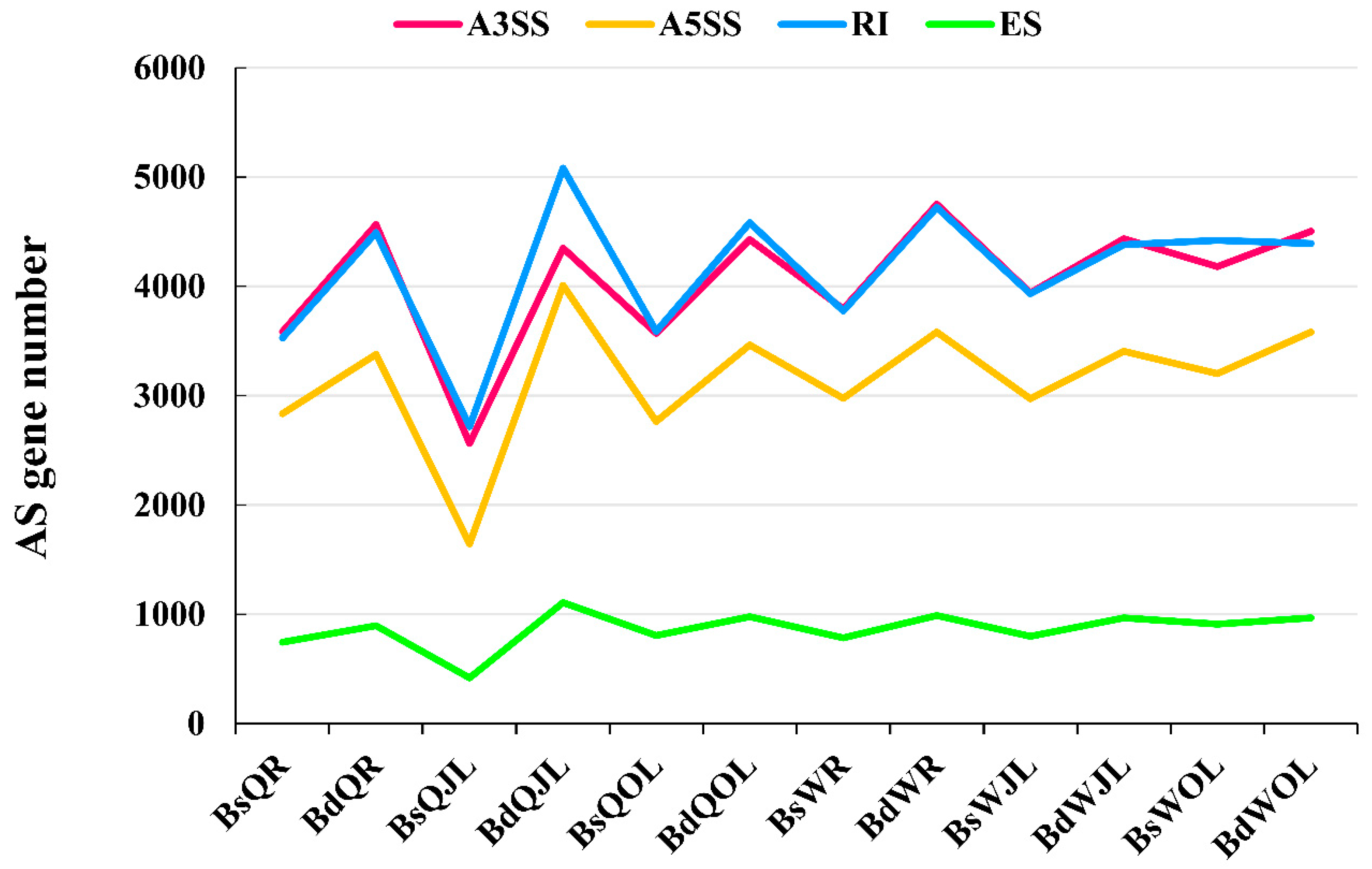
Genes | Free Full-Text | Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus | HTML
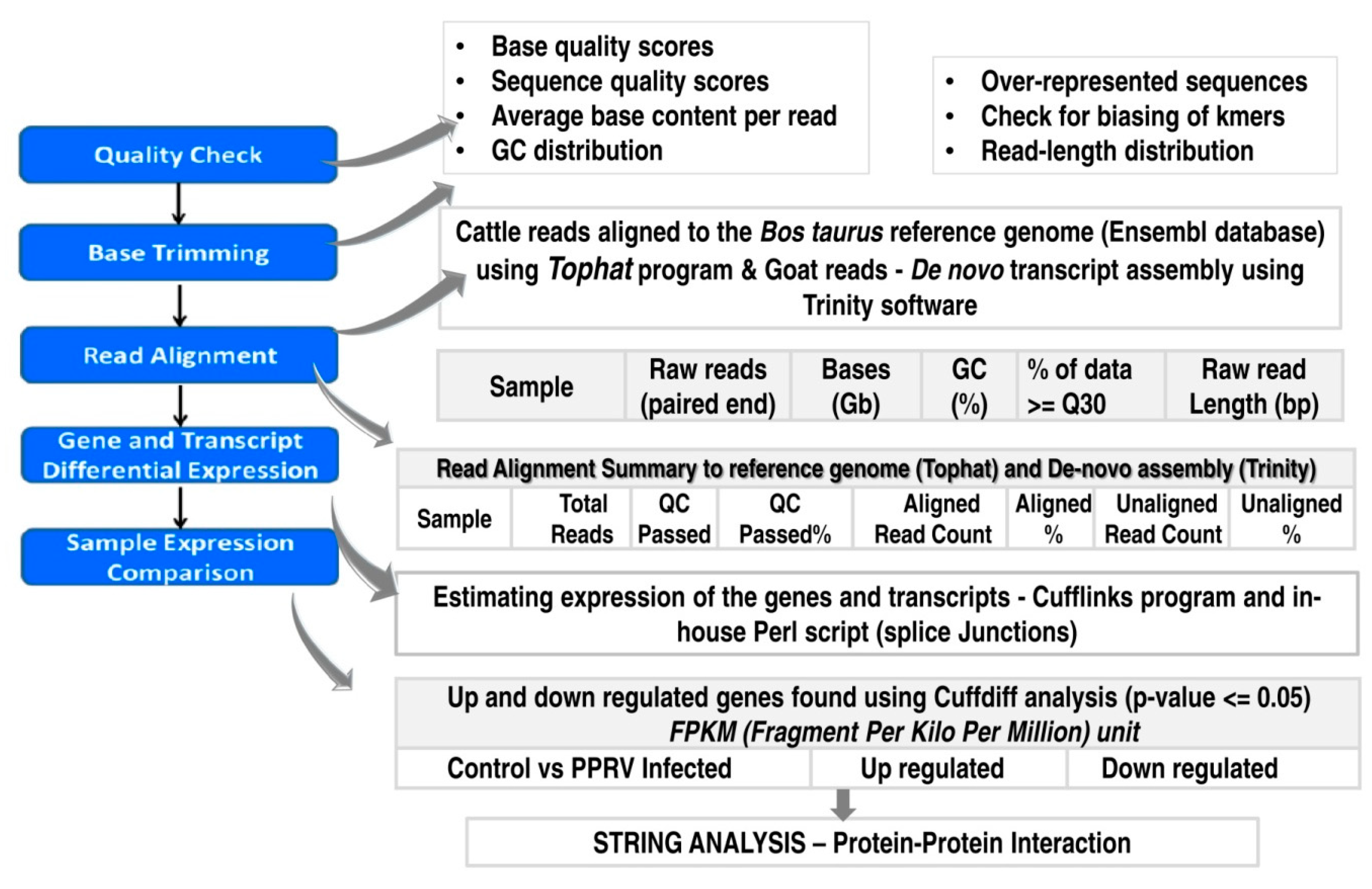
Viruses | Free Full-Text | RNAseq Reveals the Contribution of Interferon Stimulated Genes to the Increased Host Defense and Decreased PPR Viral Replication in Cattle | HTML

Genes | Free Full-Text | Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus | HTML
Spliced Leader Trapping Reveals Widespread Alternative Splicing Patterns in the Highly Dynamic Transcriptome of Trypanosoma brucei

Computational approaches for circular RNA analysis - Jakobi - 2019 - WIREs RNA - Wiley Online Library
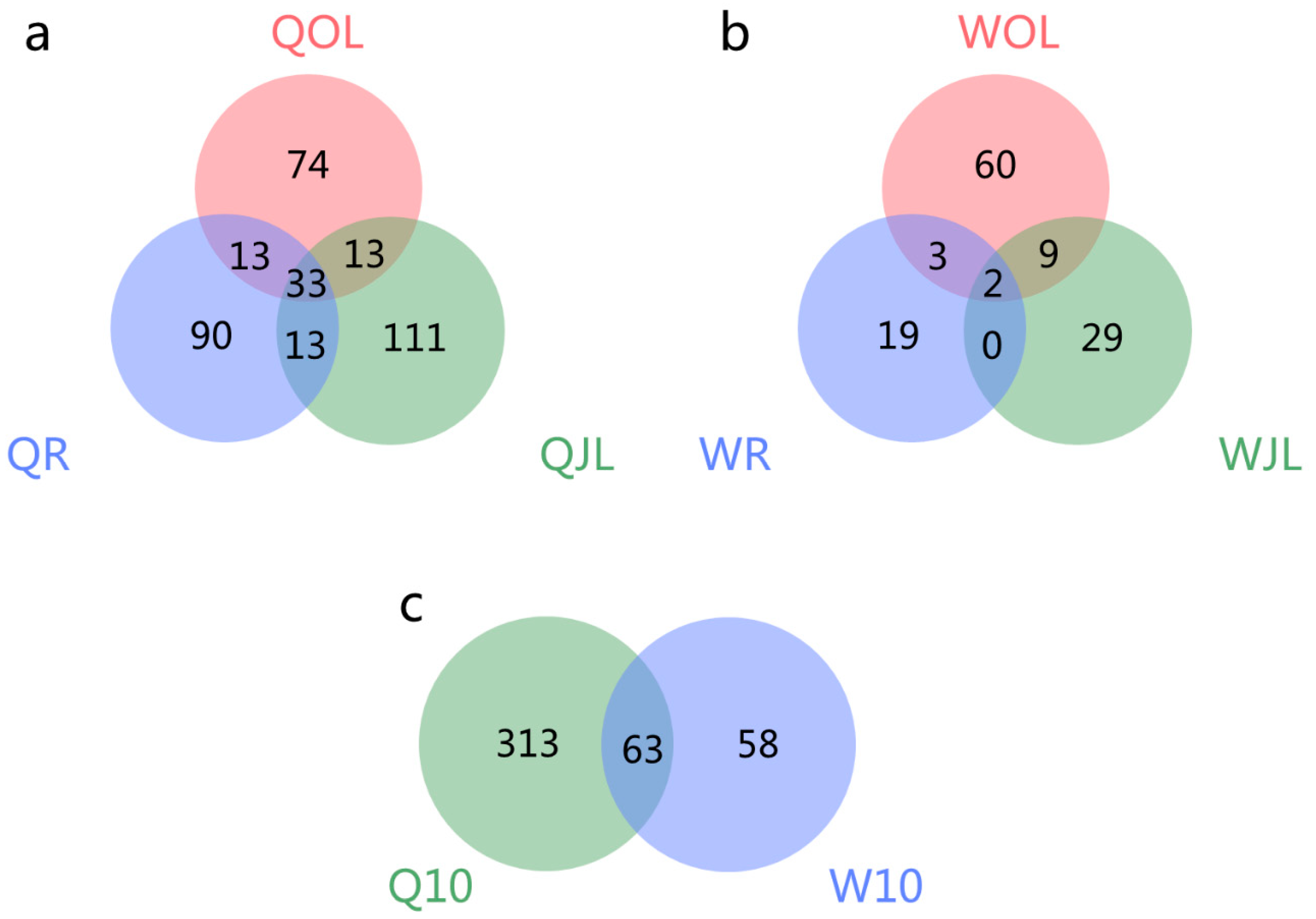
Genes | Free Full-Text | Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus | HTML
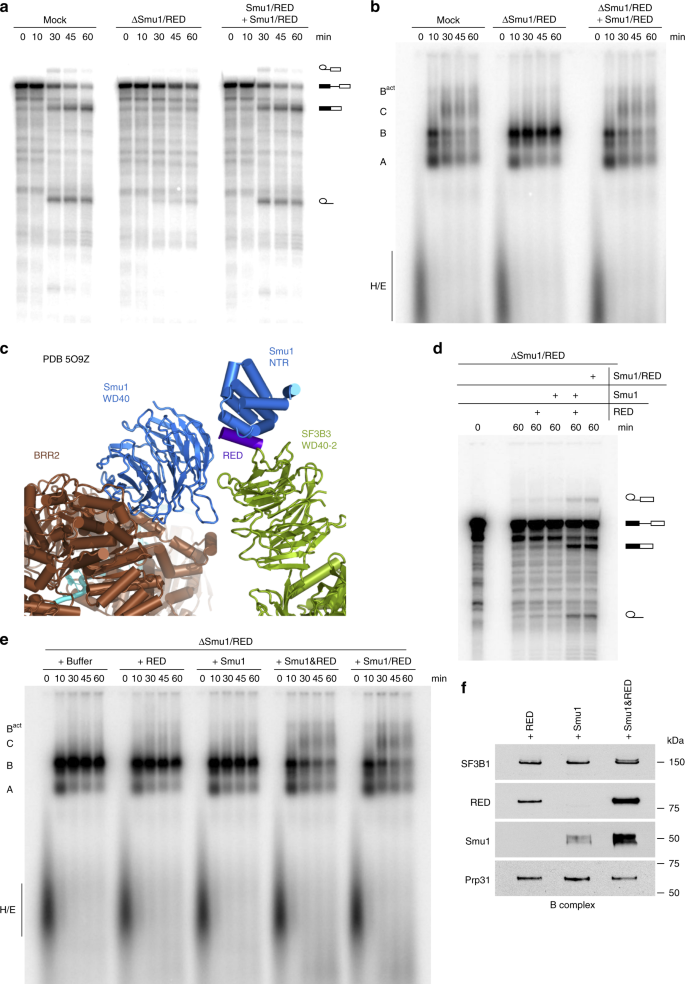
Smu1 and RED are required for activation of spliceosomal B complexes assembled on short introns | Nature Communications
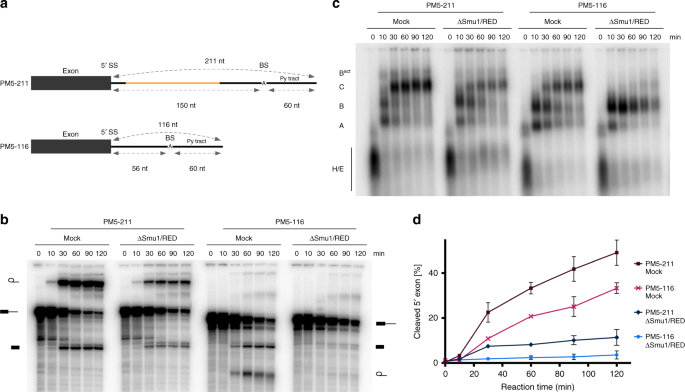
Smu1 and RED are required for activation of spliceosomal B complexes assembled on short introns | Nature Communications
PDF) Phylogenetic Comparison and Splicing Analysis of the U1 snRNP-specific Protein U1C in Eukaryotes

PDF) The Melon Zym Locus Conferring Resistance to ZYMV: High Resolution Mapping and Candidate Gene Identification
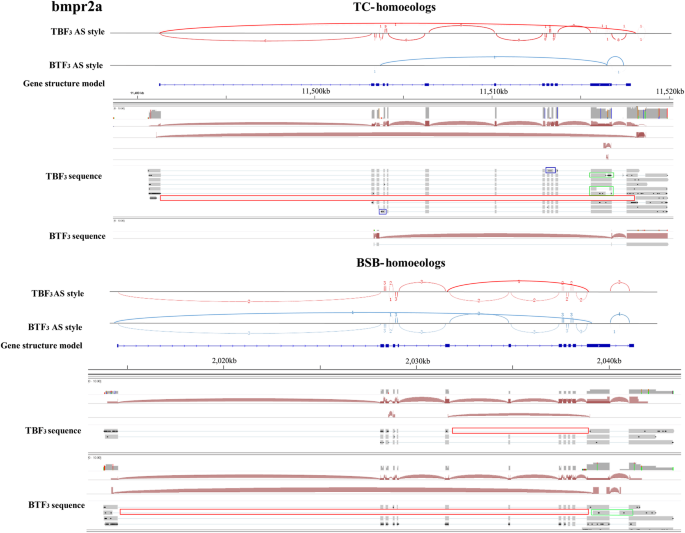
Maternal effects shape the alternative splicing of parental alleles in reciprocal cross hybrids of Megalobrama amblycephala × Culter alburnus | SpringerLink

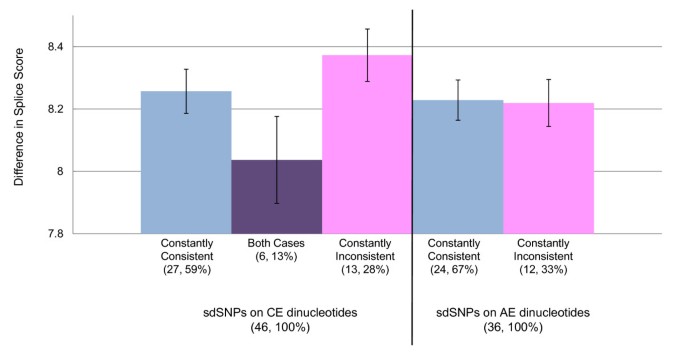
IntSplice: prediction of the splicing consequences of intronic single-nucleotide variations in the human genome | Journal of Human Genetics
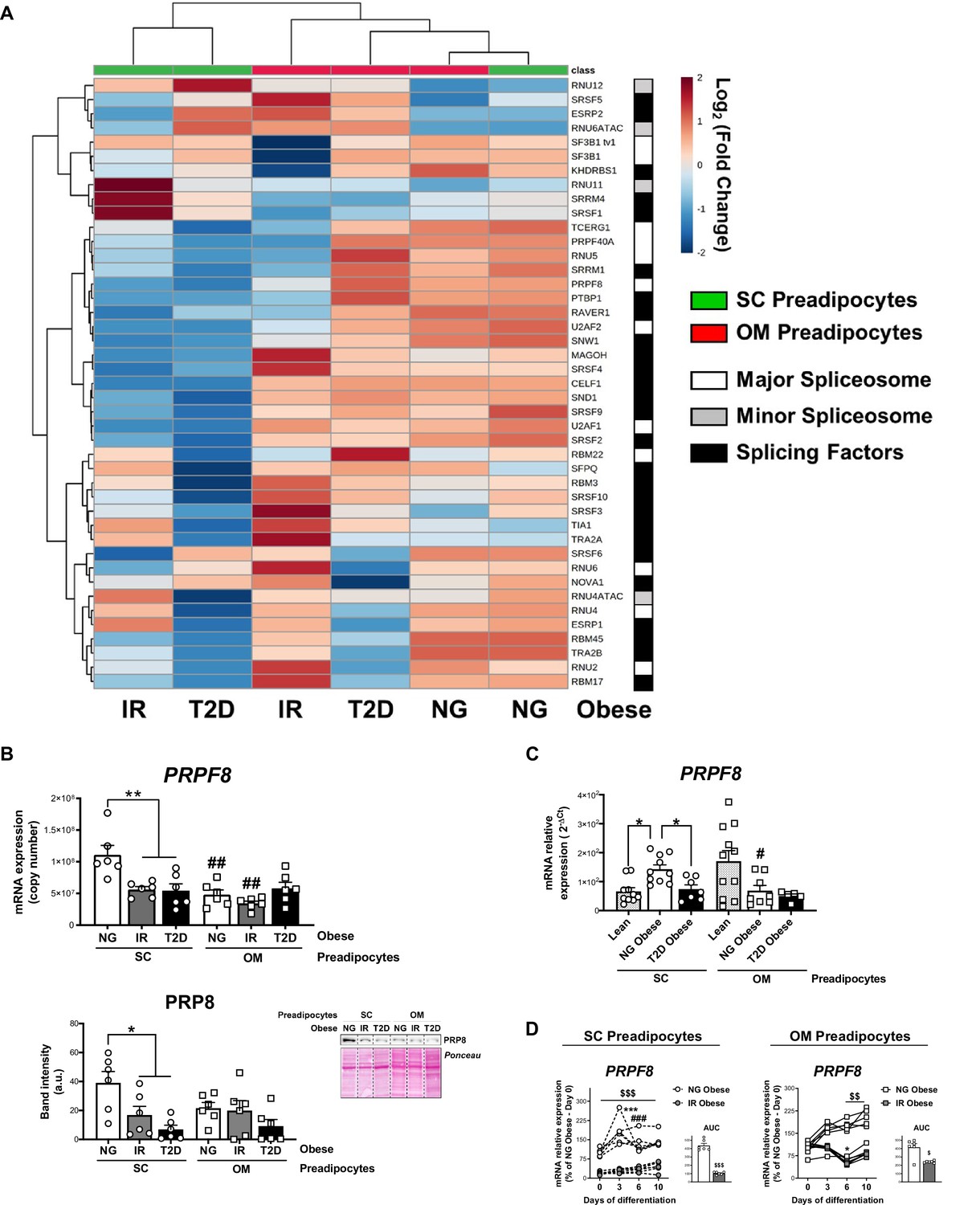
Impaired mRNA splicing and proteostasis in preadipocytes in obesity-related metabolic disease | eLife